
A
B
Exon1
sgRNA1 sgRNA2
sgRNA1 sgRNA2
Exon1 Puro
arm
arm
Exon1
*
Exon1
arm
Hygro
arm
*
HDR & Dual Selection
Cre
Exon1
*
Exon1
P4
P1
P2
P3
loxP
*
D
WT #1
bvht
dAGIL
#2
Bvht
rRNAs
WT
bvht
dAGIL
#1
bvht
dAGIL
#2
Bvht Oct4 Nanog
0.0
0.5
1.0
1.5
Relative levels of transcript
C
C C
T
G G G G
A A
C
A
G G
A
G G
A
CCTGGGGA-----------ACAGGAGGA
CCTGGGGAGGATGGATGGAACAGGAGGA
bvht
dAGIL
WT
E
#1
bvhtd
AGIL
WT
BF
DAPI
Oct-4
#2
F
0
10
20
30
WT #1 #2
bvht
dAGIL
#1 #2
bvht
H4mis
Percentage of Beating EBs
**
H
G
250
0
sgRNA-1
sgRNA-2
Exon1
d11nt
WT
bvth
dAGIL
#1
bvth
dAGIL
#2
bvth
H4mis
#1
bvth
H4mis
#2
d11nt
bvht
dAGIL
Secondary Structure
G
SHAPE Reactivity
>0.7
0.5-0.7
0.3-0.5
unlabeled <0.3
5WJ
Figure 2.
Bvht
AGIL Motif Is Necessary for Formation of Contracting EBs
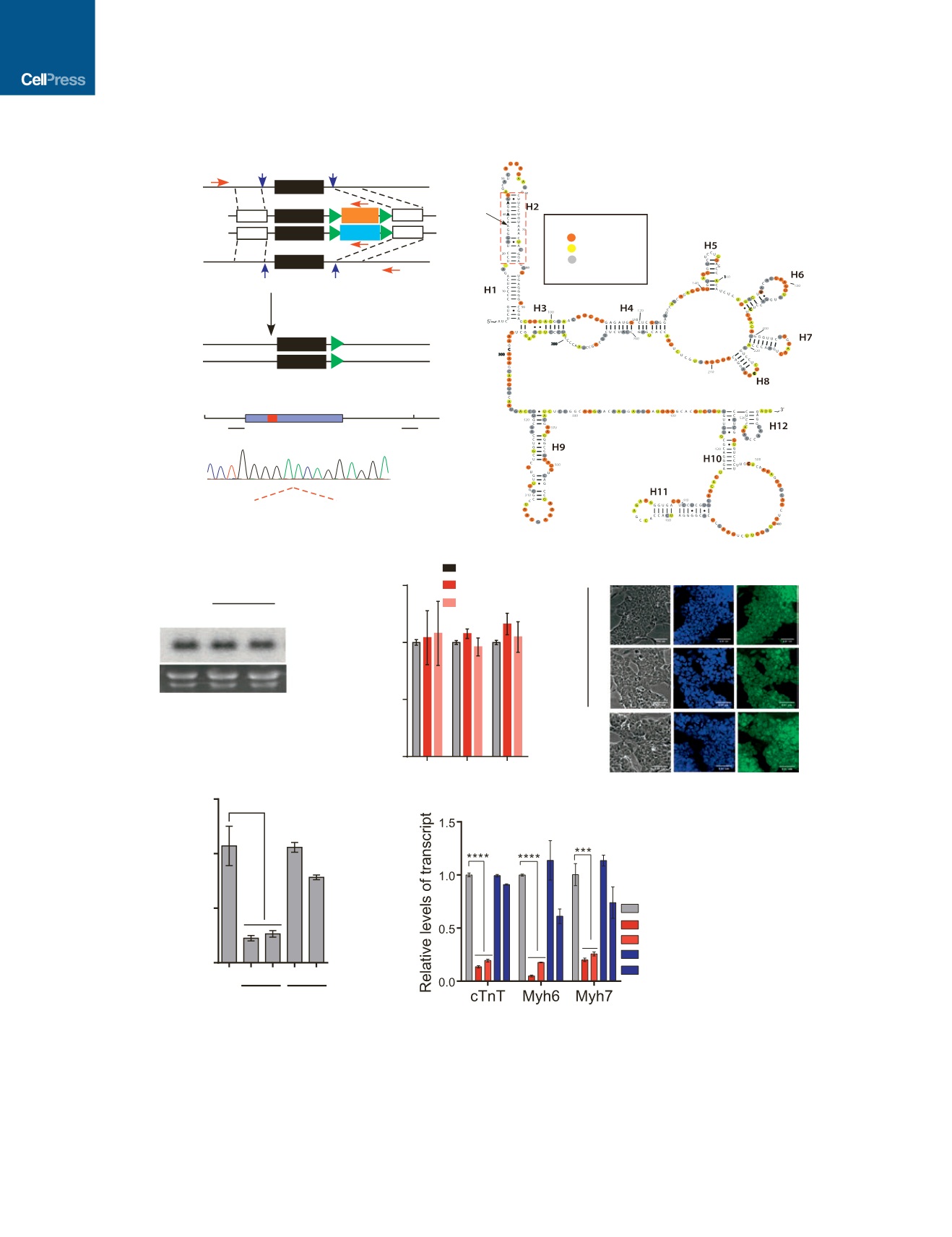
(A) Schematic showing the strategy of introducing mutations in
Bvht
endogenous locus. Two small guide RNAs (sgRNAs) and two repair templates including
different selection cassettes (Puro or Hygro) as indicated are applied for CRISPR/Cas9-mediated HDR. After dual selection, both alleles will be mutated at
designated loci. The selection cassettes are then removed by Cre recombinase-mediated recombination. Asterisk, mutations; triangle, loxP site; P1, P2, P3, and
P4 are primers for PCR-based screening.
(B) Diagram showing the positions of sgRNAs and d11nt in
Bvht
endogenous locus. Partial DNA sequencing trace of the PCR product of
bvht
dAGIL
ESC
genomic DNA.
(C) Secondary structure of
bvht
dAGIL
was derived from SHAPE probing experiment. d11nt indicates the deleted 11 nt sequences from AGIL motif.
(legend continued on next page)
40
Molecular Cell
64
, 37–50, October 6, 2016


















