
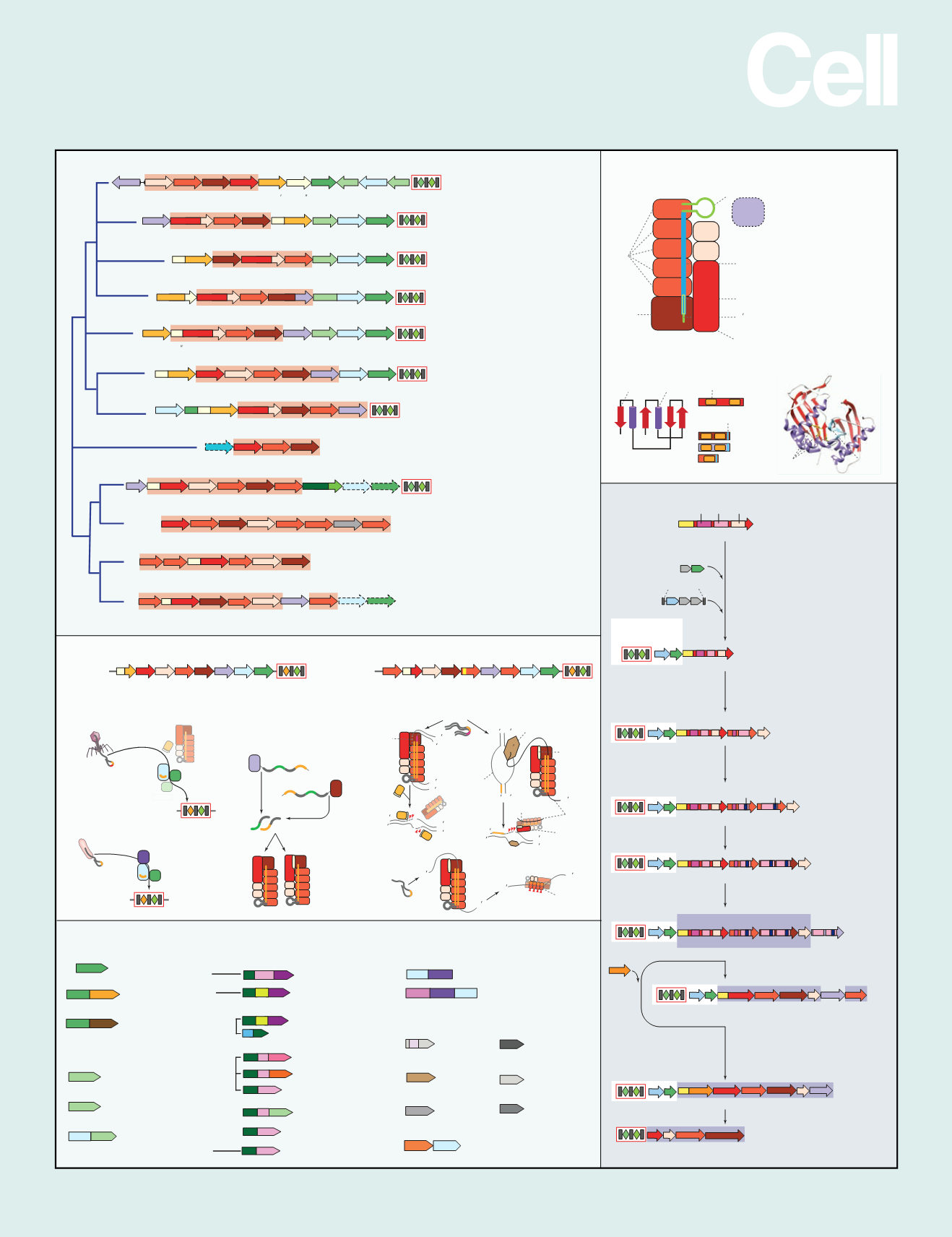
Classification
Effector complexes
Ancillary proteins
Type I
Some III-B
Type I and III
Type I and III
I-C
Generalized organization
Adaptation
crRNA maturation
and loading
I
III
IV
csy2
GSU0054
csy1
csy3
I-F
cas2
cas1
cas6f
cas5 cas7
cas3
cas8f
Yersinia pseudo-
tuberculosis
YPK_1644-YPK_1649
IV
cas7 cas5
cas8-like
csf2
csf1
csf4
csf3
dinG
Acidithiobacillus
ferrooxidans
AFE_1037-AFE_1040
DNA?
DNA
DNA
DNA
DNA
DNA
DNA
DNA
DNA
DNA
DNA?
RNA
RNA?
RNA?
RNA
I-E
cas6
cas2
cas1
cas5
cas7
cas8e
cse1
SS (cas11)
cse2
cas3
Escherichia coli
K12
ygcB-ygbF
Geobacter
sulfurreducens
GSU0051-GSU0054
GSU0057-GSU0058
cas3
I-U
cas8u2
cas2
cas1
cas7 cas5 cas6 cas4
cas3
I-C
cas8c
cas2
cas1
cas4
cas1
cas4
cas4 CRISPR
cas3
cas7
cas5
Bacillus halodurans
BH0336-BH0342
cas5
cas7
cas8a1
SS (cas11)
csa5
cas6
cas2
I-A
cas3
Archaeoglobus fulgidus
AF1859,AF1870-AF1879
I-B
cas8b1
cas2
cas6
cas1
cas4
cas5
cas7
Clostridium kluyveri
CKL_2758-CKL_2751
csc2 csc1
I-D
cas6 cas4
cas2
cas1
cas10d
cas5
cas7
Cyanothece sp.
8802
Cyan8802_0527-
Cyan8802_0520
cas7
cas7
cas5
csm6
csm5
csm4
csm3
III-A
cas6
cas2
cas1
cas10 SS (cas11)
csm2
Staphylococcus
epidermidis
SERP2463-SERP2455
III-B
cmr3 cmr4
cmr6
cas7
cas7
cas7
cmr1
cas5
cas6
cas2
cas1
cas10
SS (cas11)
cmr5
Pyrococcus furiosus
PF1131-PF1124
III-C
cas7
cmr6
cas7
cmr1
cmr4
cmr3
cas7
cas5
SS (cas11)
cmr5
cas10
Methanothermobacter
thermautotrophicus
MTH328-MTH323
III-D
cas7
cas7
cas5
csm5
csx10
csm3
cas10
SS (cas11)
csm2
cas7
csm5
cas7
csm5
Synechocystis sp.
6803
sll7067-sll7063
all1473
CRISPR
CRISPR
CRISPR
CRISPR
CRISPR
CRISPR
CRISPR
CRISPR
Origin
cas2
cas1
Terminal repeats
Toxin-antitoxin
system
Ancestral
adaptive
imminuty
Class 1
system
Effector module
Reduction of large subunit (LS)
Loss of adaptation module
and Cas6
Ancestral
Type III
system
Ancestral
Type I
system
Ancestral
Type IV
system
Duplication of Cas7
RRM1
HD
G-rich loop
G-rich loop
RRM2
Helical
cas7
cas10
cas10-like
cas10
RAMP
SS
cas10 cas7
Small
subunit
(SS)
cas5
SS
cas10 cas7
cas5 SS
SS
cas10 cas7
cas6
cas6
cas6
cas10
cas8
cas7
cas7
cas7
cas7
cas5
cas5
cas5
SS
SS
LS
PRIMPOL
family protein
CARF
(CRISPR-associated Rossmann fold)
Other families
pfam09455
pfam09659
pfam09651
pfam09670
pfam09002
HEPN
PDB: 5FSH,
Rv2818c
Mycobacterium
tuberculosis
TTHB152
Thermus thermophilus
PDB: 1XMX,
Vc1899
Vibrio Cholerae
PDB: 4EOG/2I71,
PF1127
Pyrococcus furiosus
CARF
COG1468
COG4343
Cas4, DNA nuclease
Cas1
GSU0057
Geobacter
sulfurreducens
PAE0079
Pyrobaculum
aerophilum
PAE0082
Pyrobaculum
aerophilum
Cas4a
Cas4
Cas4
6H
TM
HTH
RelE
aq_376
Aquifex aeolicus
aq_377
Aquifex
aeolicus
PDB: 2WTE,
SSO1445
Sulfolobus solfataricus
sll7062
Synechocystis sp.
sll7009
Synecho-
cystis sp.
PIN
ST0035
Sulfolobus tokodaii
MJ_1673
Methano-
caldococcus
jannaschii
VVA1548
Vibrio
vulnificus
TTE2665
Thermoan-
aerobacter
tengcongensis
PDB: 3QYF,
Sso1393
Sulfolobus solfataricus
pfam09623
Csa3
NE0113
Nitrosomonas europaea
WYL domain and HTH
Csx3, RNA nuclease
Csx16
Csx20
Ple7327_2251
Pleurocapsa sp.
Csx18
Csx15
Reverse transcriptase
(RT)
Cas1
Cas1
RT
Psta_1137
Pirellula staleyi
alr1468
Nostoc sp.
Cas6
Csx1
Csm6
Sfum_1354,Sfum_1355
Syntrophobacter
fumaroxidans
Cas1
Group 6
RAMP
Group 5
RAMP
Group 7 RAMP
RAMP
(Repeat
Associated
Mysterious
Proteins)
superfamily
Cas7
Cas7
Cas7
Cas7
Small
subunits
Cas5
Cas6
Cas8/Cas10
RRM fold
G-rich loop
G-rich
loop
Beta
strand
Alpha
helix
Repeat region (crRNA)
Spacer region
(crRNA)
Effector complex
5 handle region
(crRNA)
Seed region
(crRNA)
Mechanism of action
Large subunit
y1723
Yersinia pestis
Cas2, RNA nuclease
Cas3
SSO1404
Sulfolobus
solfataricus
Cas2
Cas2
LSEI_0356
Lactobacillus
casei
Cas2
Invading
RNA
Cas1
Cas2
Cas4
DNA
phage
RNA phage
PAM
Proto-
spacer
Proto-
spacer
Invading
DNA
Cas1
Cas2
Reverse
trancriptase
cas2
cas3
cas1
cas6
cas7
cas5
Small
subunit
Large
subunit
CRISPR
CRISPR
Priming
effect
Cas3
Effector
complex
cas6
pre-crRNA
crRNA
Cas5
pre-crRNA
Type I
effector
complex
Type III
effector
complex
Cas7
Cas10
Cas5
Cas6
Cas7
C N
1
1
1
3
3
3
2
2
2
4
4
4
RRM fold
RRM fold
effector proteins
Cas6
(Typical RAMP)
cas3
cas3
CRISPR
Adaptation
module
cas1
cas2
CRISPR
cas1
cas2
CRISPR
cas1
cas2
CRISPR
cas1
cas2
CRISPR
cas1
cas2
CRISPR
cas1
cas2
CRISPR
cas1
cas2
Cas6
pdb: 3I4H
cas3
cas3
cas3
CRISPR
Catalytic
residues
Three
distinct
groups:
Cas5
Cas6
Cas7
5
5
5
5
5
5
5
5
5
5
5
3
3
3
3
3
3
3
3
3
3
Type III
Type I
Type III
Type III
Targeting DNA
Targeting RNA
cas6
cmr4
cmr3
Small
subunit
cas10
cmr6
cmr1
cas2
cas1
CRISPR
Cas3
Invading
DNA
R-loop
Invading
RNA
Cmr4
or
Csm3
HD nuclease
HD
domain
mRNA
Cyclase
domain
GGDD
active site
RNA polymerase
ssDNA
3
Ancestral innate
immunity system
Insertion of casposon
next to cas10-like gene
CRISPR repeats evolved
from terminal repeats
of casposon
Cas1 originated from
the same casposon
Aquisition of Cas2-like toxin
from toxin-antitoxin system
Ancestral adaptive
imminuty system
Duplication of cas10-like
gene followed by its fission
Origin of cas7 family and
small subunit (SS)
Origin of G-rich loop
and duplication of
Cas7-like protein
Origin of RAMP
containing one
RRM domain
with G-rich loop
•
Origin of Cas8 by
inactivation of Cas10
Aquisition of Cas3 helicase
HD domain transfered from
Cas10 to Cas3
•
•
•
Duplication of cas5 and
origin of cas6 family
•
•
Duplication of RAMP and
origin of cas5 family
•
•
•
•
•
•
•
•
•
•
946
Cell
168
, February 23, 2017 ©2017 Elsevier Inc. DOI
http://dx.doi.org/10.1016/j.cell.2017.02.018See online version for
legends and references
SnapShot:Class1CRISPR-CasSystems
Kira S. Makarova,
1
Feng Zhang,
2,3
and Eugene V. Koonin
1
1
National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD 20894, USA;
2
Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA;
3
McGovern Institute for Brain Research,
Massachusetts Institute of Technology, Cambridge, MA 02139, USA


















