
II
A
A
A
C
C
C
U
B
B
B
V
VI
cas9
cas9
csn2
cas12a
DNA
DNA
RNA
cas4
cas9
cas2
cas1
cas4
cas2
cas1
cas1
cas2
cas1
cas2
cas1
cas2
cas1
cas2
cas1
cas4
cas12b
c2c1
c2c2
c2c3
cas12c
CRISPR
tracrRNA
cas4
cas12
cas13a
cas9
Cas9
Cas9
cas2
cas1
Cas1
Cas2
cas2
cas1
CRISPR
tracrRNA
crRNA
CRISPR
CRISPR
cas13a1
cas13a2
cas13b
Tentative
Streptococcus thermophilus
str0657-str0660
Neisseria lactamica
NLA_17660-NLA_17680
Francisella cf. novicida
FNFX1_1431-FNFX1_1428
Alicyclobacillus
acidoterrestris
N007_06525
Oleiphilus sp.
A3715_16885
Leptotrichia shahii
B031_RS0110445
Fusobacterium perfoetens
T364_RS0105110
Prevotella buccae
HMPREF6485_RS00335
Legionella pneumophila
lpp0160-lpp0163
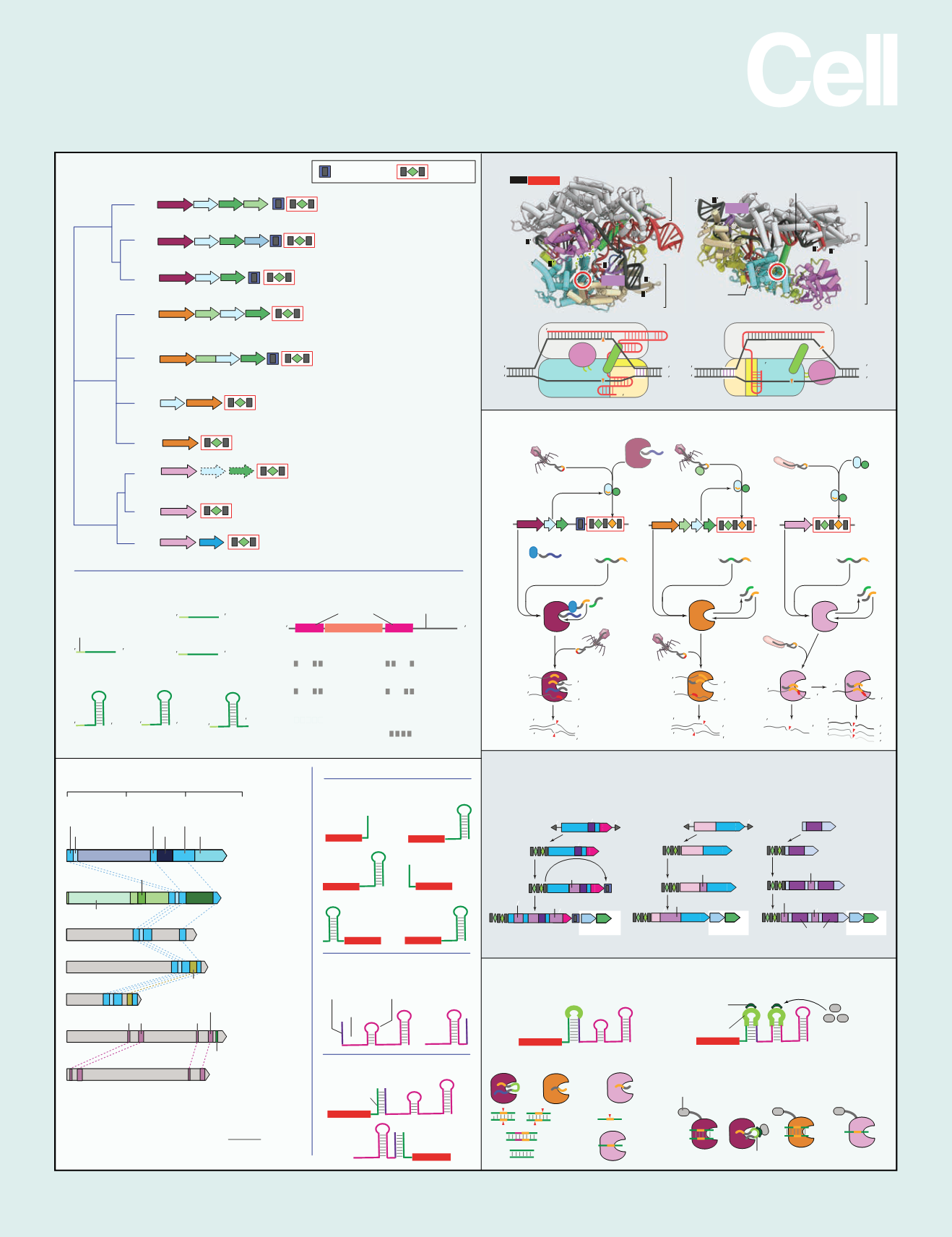
Classification
CRISPR and PAM features
RNA components
Effector protein domain organization
Effector protein structures
Cas9/
Type II
Cas12a/
Type V-A
Origin of class 2 systems
Guide RNA engineering and biotechnology applications
Recognition lobe
PAM interacting
1368
Cas9
pdb:5CZZ
Type II
Type II-A
Type II-B
Type II-C
Type V-A Type VI-A
Type II-A
Type II-AC
Type II
Type V-B
Type II
Type V-B
Type VI-A Type VI-B
Type V-A
Type V-B
>1 2 3 4 5
5 4 3 2 1<
5 4 3 2 1<
>1 2 3 4 5
U
C
Type II
Type V
Type VI
Type II-B
Type II-C
Type V-A
Type V-B
Type VI-A
Type V-A
Type V-B
Spacer
Spacer
Repeat
Type V-C
Type V-U
Type VI-A
Type VI-B
RuvC motifs
crRNA (guide RNA)
Adaptation
crRNA
maturation
Targetting,
side effects
tracrRNA
sgRNA
sgRNA for transcriptional regulation
DNA editing RNA alteration
to DNA
to RNA
Protein and small-molecule delivery
dual tracrRNA-crRNA
HEPN motifs
I II III
D
...
E
....
D
Cas12a (Cpf1)
pdb: 5B43
Cas12b (C2c1)
Cas12c (C2c3)
TnpB-like
1389
Cas13a (C2c2)
Cas13b
1500
1000
500
0
Recognition lobe
HEPN I HEPN II
R-nuclease II
HEPN II
1129
1252
1224
Protospacer adjucent
motif (PAM)
Protospacer-
flanking site
(PFS)
Target
DNA/RNA
Hairpin 1
nexus
Repeat match
AA
G
G
C
C
Hairpin 2
Cleavage
Cleavage
Site
protection
Protein or small molecule
Insertion
Deletion
knockouts
Mechanisms of action
I II
E
....
R
xxxx
H
Spacer
Repeat
Arti cial
linker
RNA aptamer
MS2 phage
coat protein
RNA aptamers
MS2 binding
regulators
Removed
upon maturation
36 nt
GAT
G
GUUU
GUU
AAC
36-48 nt
Type V-B
36-38 nt
35 nt
TS
TS
Specific insertions
Specific insertions
Specific insertions
Specific insertions
Adaptation
module
Adaptation
module
Non-autonomous
TnpB-encoding transposon
(IS605 family)
HEPN
HEPN
HEPN
HEPN
RuvC
RuvC
RuvC
RuvC
cas12
cas13
Type V
Type VI
Innate immunity
protein with HEPN
domain (a toxin?)
cas1 cas2
Adaptation
module
cas1cas2
Specific insertions
HEPN
TS
TS
HNH
Specific
insertions
tracrRNA
origin
Specific
insertions Specific insertions
Non-autonomous
IscB-encoding
transposon
RuvC
cas9
Type II
cas1 cas2
RuvC
RuvC
Conserved
bend
repeat
CRISPR
Type II-B
DNA
phage
PAM
Proto-
spacer
Proto-
spacer
Invading
DNA
Recruited
adaptation
module
Cas1 Cas2
Invading
RNA
Cas1 Cas2
Cas1 Cas2
Cas4
RNA
phage
RNA
phage
tracrRNA
Recruited host
RNAse III
Cas9
Phage
Phage
Invading
DNA
Invading
DNA
Invading
DNA
Suicidal
mode
Cas12
Cas12
Cas12
Cas9
Cas12
Cas13
pre-crRNA
Cas13
Cas13a
Cas13a
Inactivated
Cas9
cpf1
Protospacer
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
3
3
3
3
3
3
3
3
3
5
5
5
3
3
3
3
3
3
3
GUUUCA
5
3
3
3
3
3
T
C
35-
37 nt
5
3
G G
>1 2 3 4 5
G G
C
U
A
>1 2 3 4 5
>1 2 3 4 5
NN NNN
T T
5 4 3 2 1<
TT
RuvC I
RuvC II RuvC III
Nuc
HNH
Bridge helix
HEPN I
PAM interacting
Zn finger
1307
crRNA
crRNA
pre-crRNA
pre-crRNA
3
3
3
3
3
3
3
NTS TS
NTS
TS
RNA–DNA
heteroduplex
RNA–DNA
hetero-
duplex
crRNA
scaffold
sgRNA
scaffold
3′
REC lobe
NUC lobe
REC lobe
NUC
lobe
PAM
PAM
BH
WED
NTS
TS
PI
NUC lobe
REC lobe
RuvC
HNH
Nuc
Guide
BH
Guide
REC lobe
NTS
TS
PI
WED
NUC lobe
RuvC
PAM
PAM
5
5
5
3
3
328
Cell
168
, January 12, 2017 ©2017 Elsevier Inc. DOI
http://dx.doi.org/10.1016/j.cell.2016.12.038See online version for
legends and references
SnapShot:Class2CRISPR-CasSystems
Kira S. Makarova,
1
Feng Zhang,
2,3
and Eugene V. Koonin
1
1
National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD 20894, USA;
2
Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA;
3
McGovern Institute for Brain Research,
Massachusetts Institute of Technology, Cambridge, MA 02139, USA


















