
alternative PAMs by structure-based engineering of Cas9,
thereby extending genomic target range and specificity of
spCas9 (Kleinstiver et al., 2015a; Kleinstiver et al., 2015b).
Furthermore, several detailed protocols that describe the imple-
mentation of genome editing techniques in PSC systems have
been optimized and published (Blair et al., 2016; Byrne and
Church, 2015; Chiba and Hockemeyer, 2015; Yusa, 2013).
The key advantage of the CRISPR/Cas9 system over previous
systems lies in the fact that DNA-binding specificity is encoded
solely by the sgRNA and so unlike previous platforms does
not require laborious engineering of DNA binding proteins.
Thus CRISPR/Cas9-based editing has largely replaced previous
SSN technologies. Combining the cellular versatility of iPSC dif-
ferentiation with the ease of CRISPR/Cas9-mediated genome
editing proved to be a very powerful experimental approach,
and by now genome editing in hPSCs has become a standard
tool in stem cell research and human disease modeling (Johnson
and Hockemeyer, 2015; Matano et al., 2015; Schwank et al.,
2013).
One of the most exciting experiments that became possible
since the development of robust and highly efficient editing tech-
nologies in hPSCs is the genetic and functional testing of the
onslaught of empirical data generated by GWASs. Similar to
the disease-modeling approach, genome editing allows us to en-
gineer variant alleles observed in these studies found to be asso-
ciated with a specific disease in an otherwise isogeneic cellular
setting. Phenotypic comparison of such cells can reveal how
non-coding mutations, enhancer polymorphisms, and balancer
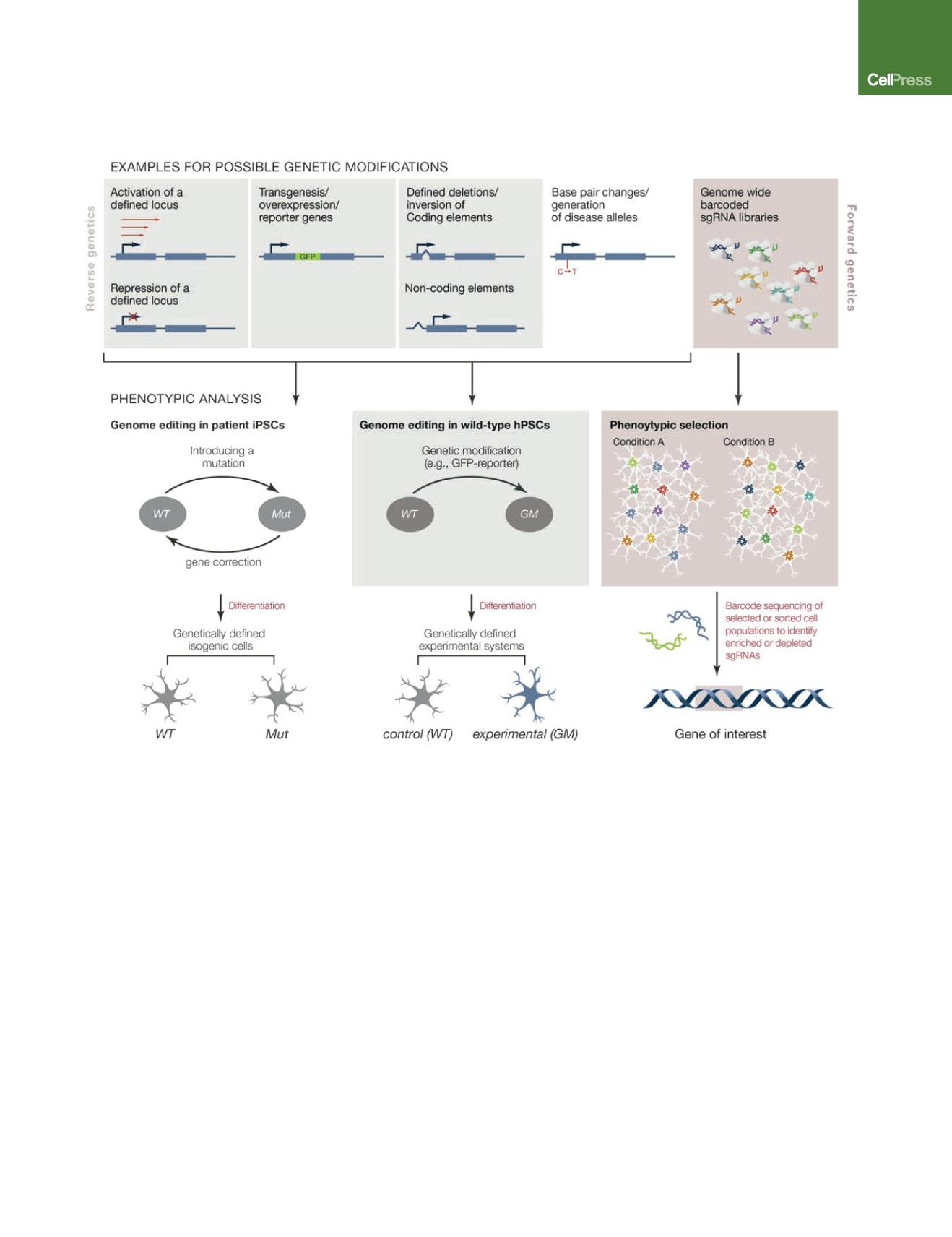
Figure 2. Genome Editing Applications in hiPSCs
Genome editing allows the genetic modification of hiPSCs. The top panel (left side) depicts examples of reverse genetic approaches to study hPSCs using
genome editing. Gene expression can be modulated (activated or repressed: CRISPRi and CRISPRa) by reversibly targeting their endogenous promoter. Genes
can be inserted to generate reporter genes or to achieve ectopic expression. Genetic information can be deleted or inverted and modifications as small as single
base pair changes can be generated to introduce mutations or polymorphisms or to repair disease-relevant mutations. The resulting genetically engineered
hPSCs differ from wild-type cells exclusively at the edited locus and are otherwise isogenic (bottom left). Parallel differentiation of these isogenic cell lines into
disease-relevant cell-types can provide the basis for the phenotypic analysis of disease-specific cellular pathologies. Phenotypes found in these cells can be
directly attributed to genetic manipulation. In addition, forward genetic approaches to study hPSCs (top right panel) became available with the development of
genome editing as a screening tool. Bulk transduction of hPSCs with either Cas9 or dCas9 in combination with genome-wide barcoded sgRNA libraries—
‘‘CRISPR cutting, CRISPRi, and CRISPRa’’—can be used to identify genes whose loss or gain of function changes the cellular representation within the infected
cell pool. Enrichment or depletion of sgRNAs can be determined by sequencing the sgRNAs, yielding candidate genes of interest (bottom right panel).
Cell Stem Cell
18
, May 5, 2016
577
Cell Stem Cell
Review


















