
a GWAS-identified risk regulatory allele on the target gene (or
genes) would be predicted to be subtler than would be expected
for monogenic diseases as discussed above. Thus, it would be
impossible to compare the disease-specific cells to a suitable
control cell line because any control cells would have a different
genetic background, which will affect the differentiation potential
of the cells and thus would prevent a meaningful comparison.
Thus, amajor challenge for using iPSCs for the study of sporadic
diseases is how to generate pairs of isogenic cells that differ at one
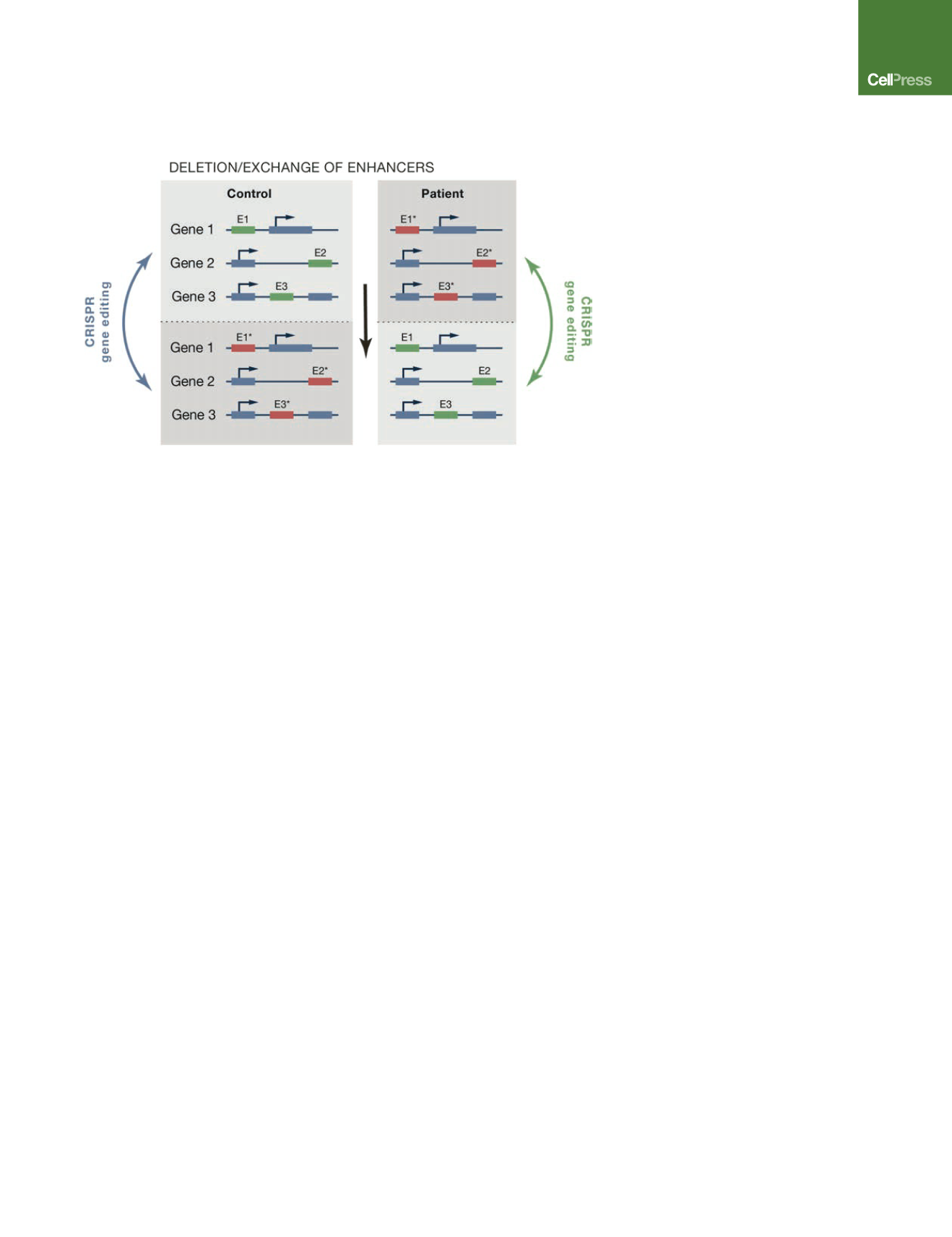
or multiple risk alleles. Figure 3 outlines a possible strategy of how
the CRISPR/Cas9 gene editing approach could be used to
generate isogenic cells that differ at multiple risk loci and thus
would enable the mechanistic study of polygenic diseases. This
approach was recently used to decipher the impact of PD-associ-
ated risk variants. Genetic engineering of a common PD-associ-
ated risk variant in a non-coding distal enhancer resulted in
deregulation of SNCA expression, a key gene implicated in the
pathogenesis of PD, by as little as 10% (Soldner et al., 2016). In or-
der to detect such subtle gene expression differences, an allele-
specific assay was developed that allowed the analysis of
cis
-act-
ing effects of candidate variants onallele-specific gene expression
as a consequence of deletion or exchange of disease-associated
regulatory elements. Detailed analysis of isogenic cells with and
without the risk allele further demonstrated that a single base
pair change causes loss of transcription factor-binding sites for
the transcription factors that otherwise function as a suppressor
of SNCA transcription on a non-risk-associated allele.
Epidemiology and population genetics suggest that Sporadic
Alzheimer Disease (SAD) results from complex interactions be-
tween genetic risk variants and environmental factors. In another
approach to study risk alleles, patient-derived hiPSCs were used
to dissect the effect of common SAD-associated non-coding
genetic variants in the 5
0
region of the
SORL1
(sortilin-related
receptor, L(DLR class) A repeats containing) gene involved in
intracellular vesicular trafficking (Young et al., 2015). While initial
experiments did not identify a consistent correlation between
SORL1 expression and either disease status or risk haplotype,
a small but significant correlation between the SAD-associ-
ated
SORL1
haplotype and the BDNF-dependent response of
SORL1 expression was found.
Figure 3. Strategy to Generate Isogenic
iPSCs that Differ at Multiple Risk Alleles
GWASs have identified genomic loci that may
slightly increase the risk of developing a sporadic
disease. The key challenge of using patient-derived
iPSCs to get mechanistic insight into risk alleles is
to create meaningful control cells. CRISPR/Cas9-
mediated gene editing would allow exchanging risk
(red squares) and protective (green squares) alleles
and generating appropriate control cells that differ
exclusively at the risk loci under study.
Nuclease Specificity and Off-Target
Considerations
SSNs are enzymes that are targeted to
specific sites in the genome, but their spec-
ificity can vary and promiscuous binding
to so called off-target sites can lead to
unwanted cutting and modifications. Stra-
tegies to predict, identify, and reduce these off-target events are
largely dependent on the SSN design, organism, and cell type
and have already been to some extent implemented in hPSCs.
Understanding the frequency and impact of off-targets is highly
relevant to the development of SSNs for clinical applications and
their reliable use in basic research (Gabriel et al., 2011).
Several studies recently addressed the specificity of Cas9 and
its off-target action (reviewed in Wu et al., 2014a). Genome-wide
binding studies of dCas9 expressed in mESCs demonstrated
that Cas9 can associate with a large number of genomic sites,
but off-target cutting of the catalytically active Cas9 at a subset
of these sites was infrequent (Wu et al., 2014b). Similarly, single-
molecule imaging of Cas9 in living cells has demonstrated that
Cas9 searches for target sites by 3D diffusion, and that, in
contrast to on-target events, off-target binding events are, on
average, short-lived (<1 s) (Knight et al., 2015).
While these data argue for the high specificity of Cas9, data in
cancer cells suggest that off-targets can be frequently detected
(Frock et al., 2015; Fu et al., 2014; Tsai et al., 2015; Wang et al.,
2015b). For example, when using GUIDE-seq (Tsai et al., 2015), a
protocol optimized in U2OS and HEK293 to detect off-targets
more reliably than other methods such as ChIP-seq, Tsai et al.
found many off-targets that computational algorithms had failed
to predict. Based on these datasets Tsai et al. proposed that
shorter guide sequences that only have about 17-nt homology
to the target sequence would improve specificity (Fu et al.,
2014). Moreover, the GUIDE-seq protocol was also used to en-
gineer CRISPR-Cas9 nucleases with altered PAM specificities
(Kleinstiver et al., 2015a, 2015b) and reduced off-targets (Klein-
stiver et al., 2016).
An alternative protocol called BLES-seq, based on directly
labeling the DSBs generated by the nuclease in situ followed
by enrichment through streptavidin affinity purification and
next-generation sequencing (Crosetto et al., 2013), was origi-
nally developed to detect DSBs caused by replicative stress
by stalled replication in HeLa cells and mouse B lymphocytes.
This protocol was further developed to assess Cas9 off-target
frequencies of Cas9 and to rationally engineer Cas9 nucleases
with improved specificity (Ran et al., 2015; Slaymaker et al.,
2016).
Cell Stem Cell
18
, May 5, 2016
579
Cell Stem Cell
Review


















