
domain via a multimeric peptide array (SunTag), wherein each peptide domain could bind to a
single-chain variable fragment (scFv) fused to VP64 [53]; fusing dCas9 to a synergistic tripartite
activator system containing VP64, the activation domain of p65 (p65AD), and Epstein-Barr virus
R transactivator (Rta) [54]; and combining dCas9-VP64 with a modi
fi
ed sgRNA engineered with
two copies of an MS2 RNA hairpin that could recruit p65AD and the human heat shock factor 1
(HSF1) activation domain via interaction with the MS2-binding protein [48]. A systematic
comparison of the ef
fi
cacy of these methods revealed that these systems perform comparably
but are dependent on the genomic and cellular context [55], suggesting that activation ef
fi
ciency
varies for different genes and in different types of cell. In the future, simpler, yet more effective,
tools for RNA-guided gene activation should be further developed.
To repurpose more complex gene regulation, sgRNA was engineered as a class of
‘
scaffold
’
RNAs (scRNAs) that directly recruit transcription effectors without protein fusion [56]. scRNAs
are generated by fusing RNA hairpins to the sgRNA, which interact with the cognate protein to
recruit activators or repressors. Using engineered scRNAs, multiple genes can be simulta-
neously activated and repressed in the same cells. In addition to using scRNAs, multiple
orthogonal species of dCas9s could also provide a platform for complex transcription regulation
and sophisticated manipulation of the transcriptome.
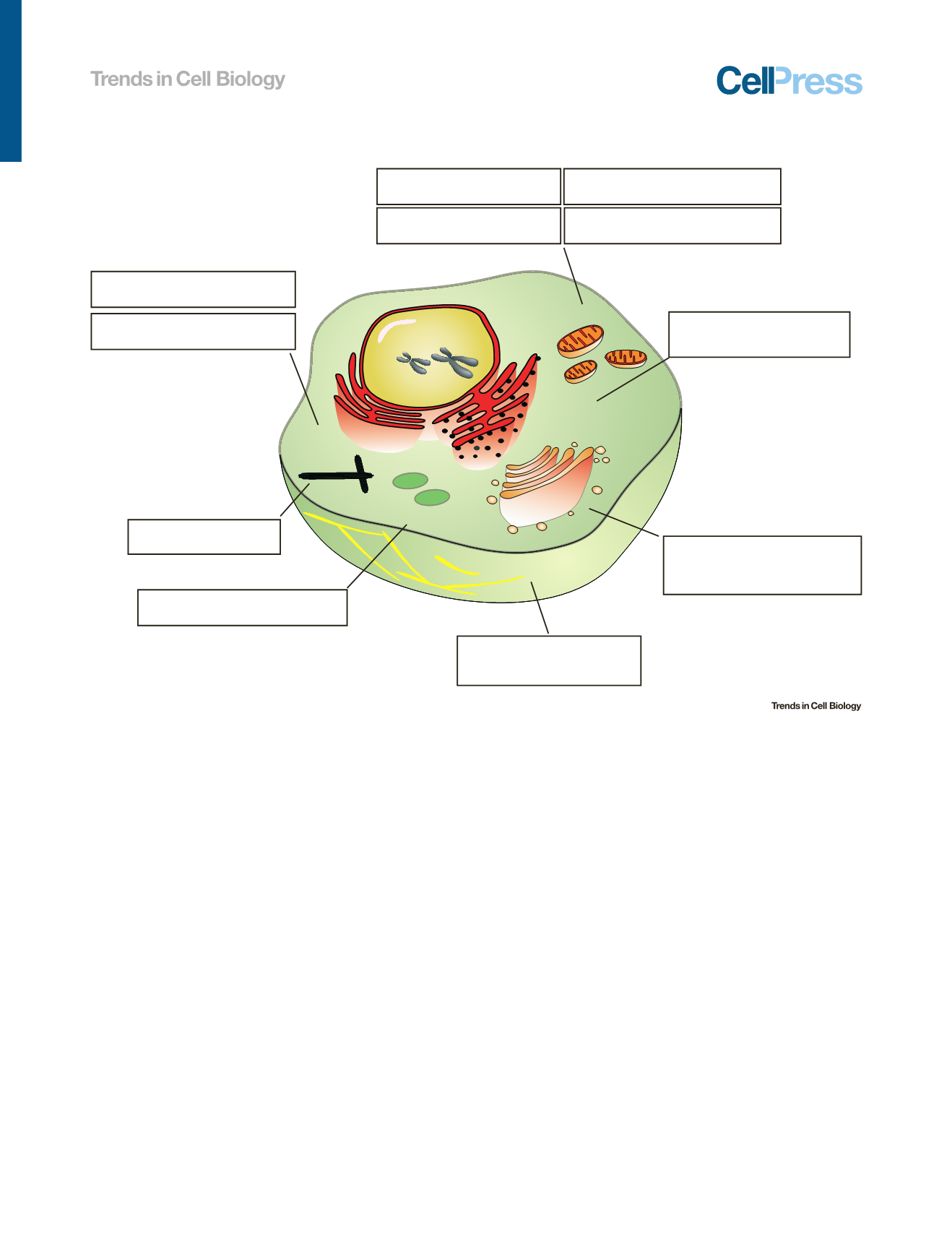
Mitochondrion
Golgi apparatus
Lysosome
Endoplasmic reƟculum
(ER)
Nucleus
Microtubule
Chromosome
Knockout screen to idenƟfy genes for cell
growth suppression due to inhibiƟon of ETC
DeleƟng a linker region in CP190
altered spindle morphology and
led to DNA segregaƟon errors
∆
Ire1
α
reduced cleavage of ER-
targeted mRNA
∆
Asp caused spindle defects
in neuroblasts
∆
RPS25 + transgene RPS25-SNAP
revealed the kineƟcs of the 40S
subunit recruitment to HCV IRES
Genome-wide screen to idenƟfy genes
required for
S. aureus
toxin
α
HL
suscepƟbility in human myeloid cells
idenƟfied roles for new proteins
∆
ATP7A damages the mitochondrial
redox balance
∆
FASTKD2 caused defecƟve processing
and expression of mitochondrial RNA
Repair of the ARID5B moƟf of
rs1421085 restored thermogenesis
∆
ATF4 revealed ATF4 binds and acƟvates
NLRP1 promoter during ER stress
∆
NPC1 revealed NPC1 moves cholesterol
across the lysosomal glycocalyx
Ribosome
Centrioles
Figure 3. Examples of Applying Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-Associated Protein 9 (Cas9)
Technology to Study Cellular Organelles.
The
fi
gure illustrates exemplar studies in particular organelles, with more details listed in Table 1 (main text).
880
Trends in Cell Biology, November 2016, Vol. 26, No. 11


















