
for dCas9-KRAB expression; Figure S5A). There was no
silencing of the TetO promoter in low-passage and high-passage
iPSCs, suggesting that long-term culturing (>3 months) does not
cause silencing. However, cardiac progenitors (day 5) and iPS-
CMs (day 15) lose 20% and 50%–80% of the doxycycline
response, respectively. Prolonging the duration of doxycycline
treatment (from 2 to 7 days) and splitting the cells improved
doxycycline response (as measured by mCherry expression)
in iPS-CMs (Figure S6C). For this reason, we initiated all of
our knockdowns on day 5 post-differentiation to obtain the
maximum amount of target gene silencing. It is worth noting
that with CRISPRi, only minute amounts of the dCas9-KRAB pro-
tein are necessary to induce a knockdown. Hence, knockdown
might occur even in cells that do not show detectable mCherry
expression (Figure S5).
The knockdown of the
HERG
potassium channel in iPSCs was
highly efficient (>95%), while in iPS-CMs it was only 60% effec-
tive. We hypothesize that the reduction in the efficiency of
HERG
knockdown is partially due to activation of other
HERG
isoforms
in iPS-CMs. We further investigated whether knocking down the
HERG
potassium channel in iPS-CMs would recapitulate a phys-
iologically relevant cellular phenotype. We found that knocking
down
HERG
in iPS-CMs lead to a prolonged beat duration and
the appearance of a shoulder during the downstroke, as
measured using the GCaMP signal (which can be used as a sur-
rogate for the action potential) (Huebsch et al., 2015) (Figures 6D
and 6E). We confirmed the prolongation of action potential dura-
tion by patch-clamp electrophysiology in the
HERG
knockdown
samples (Figures 6F). We expected this result, because the
HERG
potassium channel pumps potassium ions out of cells
to lower the inner membrane potential during diastole. This
cellular phenotype recapitulates aspects of the phenotype
observed in LQT patients and their iPS-CMs (Schwartz et al.,
2012; Spencer et al., 2014).
DISCUSSION
In this study, we combined the power of human iPSC technol-
ogy, which generates functional human cells, with inducible
CRISPR-based genome editing and modulation technologies.
Using the TetO inducible system, we deploy the newly devel-
oped CRISPRi system in the AAVS1 safe–harbor locus of human
iPSCs to enable precise control of transcript silencing upon addi-
tion of doxycycline. With this approach, we rapidly and efficiently
generated loss-of-function phenotypes in iPSCs and their cell-
type derivatives to study mechanisms in development and
disease. We introduced a single doxycycline-inducible vector
system into the AAVS1 safe-harbor locus to gain tight transcrip-
tional control of dCas9-KRAB (for CRISPRi) and Cas9 (for
CRISPRn) for gene knockdown and knockout studies, respec-
tively. This inducible vector system helped us precisely control
the timing of knocking down the expression of target genes in
a clonal iPSC line carrying the gRNA of interest. We were also
able to efficiently target the CRISPRi vector into non-iPSC hu-
man cells (T-lymphocytes) and show efficient levels of transgene
knockdown, which demonstrates the versatility of using the
CRISPRi system in a wide range of cell types. This system
can be readily targeted to other human cellular models in vitro
and also to mouse models (Soriano, 1999) by exchanging the
AAVS1-homology arms with the ROSA26-specific knockin arms.
We found that in iPSC populations, CRISPRi produced a ho-
mogeneous and rapid loss-of-function phenotype compared
to CRISPRn. CRISPRi avoids potential complications associated
with incomplete loss-of-function and gain-of-function pheno-
types in cell populations produced by Cas9-induced hypomor-
phic alleles. Therefore, CRISPRi represents a powerful tech-
nology for repressing gene expression in bulk populations and
especially when performing genome-scale phenotypic screens.
Every CRISPRi iPSC that contained a target-specific gRNA
OCT4
NANOG
SOX2
T
PAX6
OCT4
Knockdown
OCT4
NANOG
SOX2
T
PAX6
NANOG
Knockdown
OCT4
NANOG
SOX2
T
PAX6
SOX2
Knockdown
Maximal
mRNA Expression
100%
75%
50%
25%
0%
– Dox 1
2
3
4
5
6
7
Days on Dox
A
+ Dox (RPM)
– Dox (RPM)
Activation
Repression
dCas9-KRAB
GCaMP
VIM
~ 30 fold
repression
B
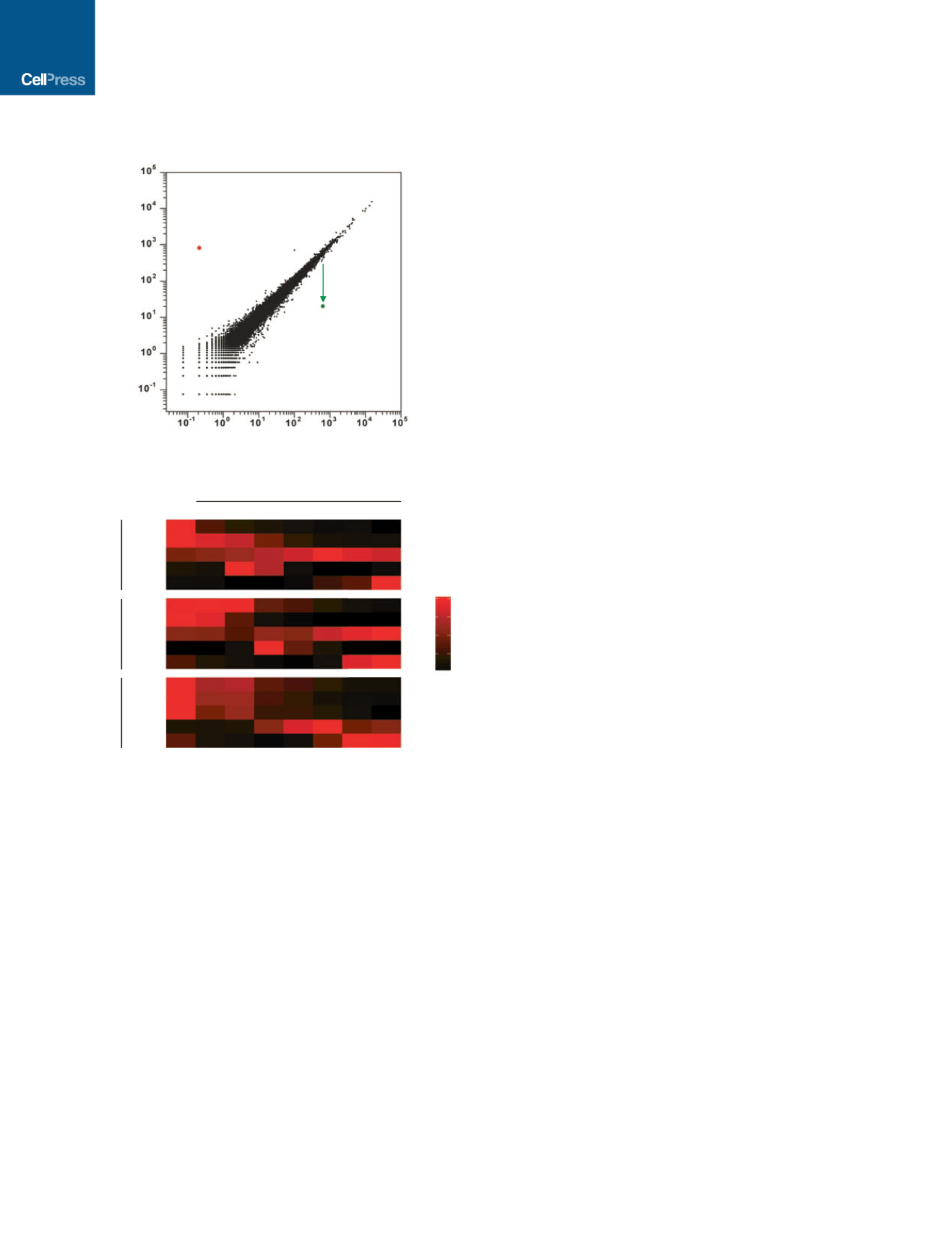
Figure 5. RNA-Seq and TaqMan qPCR Analysis
(A) RNA-sequencing RPMs (reads per million) are plotted for CRISPRi cells
stably expressing a gRNA targeting the GCaMP transgene (GCaMP g+56)
cultured in the absence or presence of doxycycline. CRISPRi knockdown
is specific to the GCaMP transcript, and few off-target transcriptional
changes were observed. Data represent two independent biological
replicates.
(B) Heatmap of TaqMan qPCR of stable clones containing a single gRNA
against the gene of interest (
OCT4
,
NANOG
, and
SOX2
) as a function of days
after doxycycline treatment. Analysis shows that by day 3, over 80% of the
target transcript is depleted. Three housekeeping genes (
18S
,
GAPDH
, and
UBC
) were used to measure relative transcript levels. Each data point is an
average of two to four technical replicates. TaqMan probes are listed in
Supplemental Experimental Procedures.
548
Cell Stem Cell
18
, 541–553, April 7, 2016
ª
2016 Elsevier Inc.


















