
loss of transcript expression and rapid cell differentiation when
targeting
NANOG
and
OCT4
within 5–7 days of knockdown initi-
ation. With CRISPRn, even after 2 weeks of doxycycline treat-
ment, a significant fraction (30%–40%) of the cells remained
NANOG and OCT4 positive and maintained their pluripotency.
Therefore, we focused on using CRISPRi as a loss-of-function
tool in subsequent experiments.
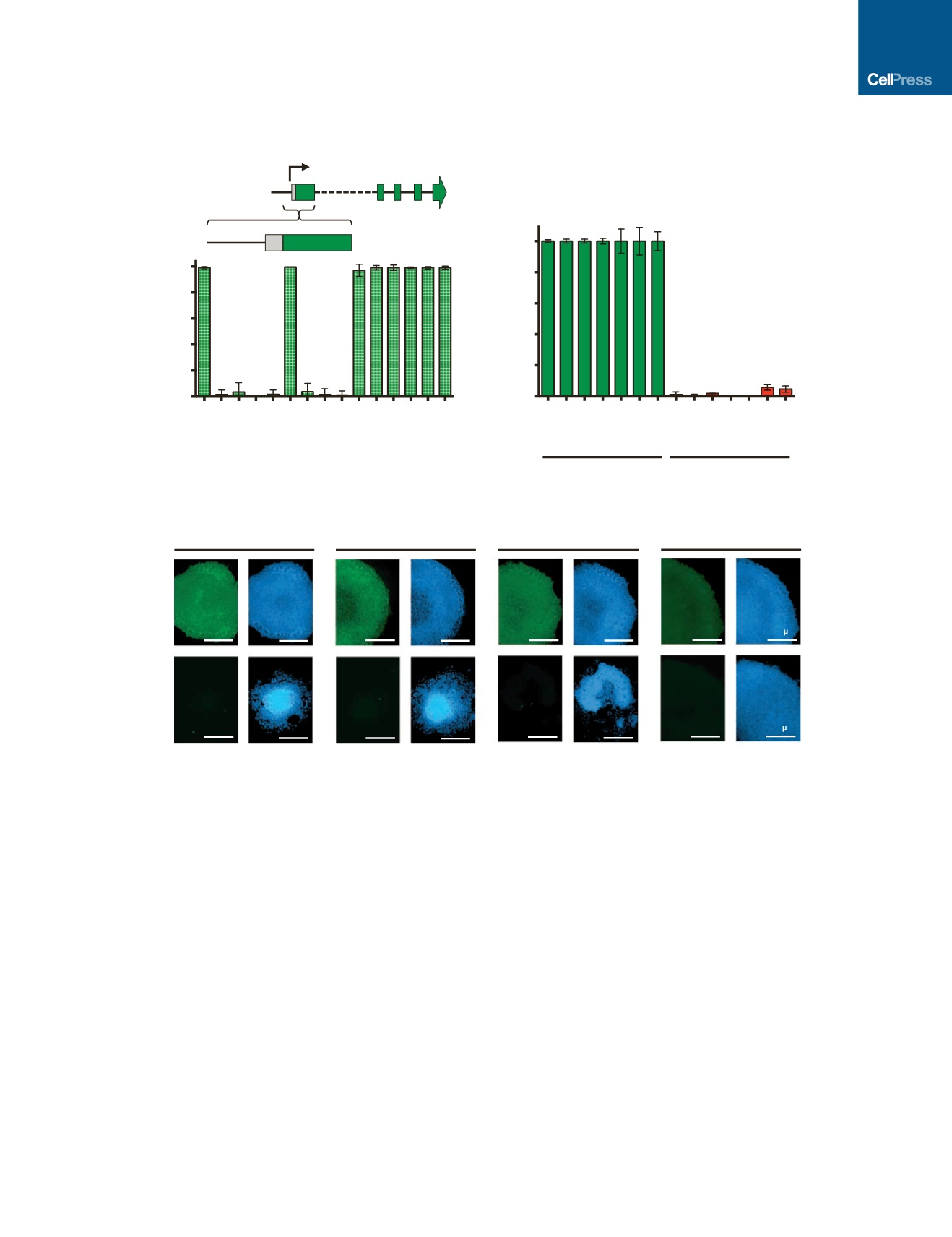
CRISPRi Is Most Effective near the TSS
To further test the efficacy of gRNAs in CRISPRi, we designed
multiple gRNAs that target near the TSS of OCT4. With flow
cytometry assays for OCT4 staining (Figure 3A), we found that
most gRNAs targeting near the TSS (approximately 150 bp
to +150 bp around the TSS in this study) were highly effective at
gene knockdown, but gRNAs targeting significantly (>700 bp)
downstream of the TSS were not. This result agrees with previous
data (Gilbert et al., 2014) and suggests that CRISPRi primarily
blocks transcription at initiation, which reduces the likelihood
of off-target effects from transcript interference elsewhere in
the genome. Following these design criteria, for subsequent
gene targets, we designed gRNAs to target near the TSS.
CRISPRi Efficiently Knocks Down a Broad Range of
Genetic Loci
To test the efficiency of CRISPRi across a broad range of genetic
loci in both iPSCs and differentiating/differentiated cell types, we
5 4
3
2
1
Transcription Start Site
1
OCT4
Locus
(Not to scale)
– Dox
– 142 (T)
– 105 (NT)
– 7 (T)
+ 22 (NT)
+ 42 (T)
+56 (T)
+104 (NT)
+126 (T)
+701 (T)
+1305 (NT)
+2390 (NT)
+3410 (NT)
+4580 (T)
+5632 (T)
0
20
40
60
80
100
Position Relative to TSS (bp)
% of OCT4
+
Cells
C
B
OCT4 –
NANOG –
SOX2 –
BAG3 –
ROCK1 –
GSK3b –
HERG –
OCT4 +
NANOG +
SOX2 +
BAG3 +
ROCK1 +
GSK3b +
HERG +
0
20
40
60
80
100
– Dox
+ Dox
A
OCT4
gRNA
– Dox
OCT4
NANOG
OCT4
DAPI
DAPI
DAPI
DAPI
DAPI
DAPI
SOX2
NANOG
SOX2
DAPI
DAPI
BAG3
BAG3
500 m
500 m
+ Dox
NANOG
gRNA
SOX2
gRNA
BAG3
gRNA
% of Maximal RNA Expression
Figure 3. CRISPRi Knockdown Is Efficient in iPSCs
(A) Efficiency of gRNA knockdown based on proximity to the transcription start site (TSS). The binding location of each gRNA is indicated relative to the TSS of the
OCT4
locus and whether it targets the template (T) or non-template (NT) strand. Only gRNAs targeting near the TSS (approximately ±150 bp) effectively knocked
down
OCT4
.
(B) TaqMan qPCR analysis of stable iPSCs containing gRNA against the gene of interest showed greater than 90% knockdown efficiency after 7 days of
doxycycline induction in different endogenous genetic loci.
(C) Immunostaining of stable clones containing a single gRNA against the gene of interest (
OCT4, SOX2
,
NANOG,
and
BAG3
). After 7 days of doxycycline
treatment, there was a complete knockdown of the protein of interest (green). As expected, DAPI staining revealed that knocking down
OCT4
,
NANOG
and
SOX2
resulted in loss of pluripotency and clear morphological changes. Also, knocking down
BAG3
did not cause a loss of pluripotent morphology, as indicated
by the distinct and round colony edges.
Error bar represents SD.
Cell Stem Cell
18
, 541–553, April 7, 2016
ª
2016 Elsevier Inc.
545


















