
indicated by a clear loss of pluripotent cell morphology (Hayashi
et al., 2015). In general, Cas9 can disrupt gene function at any
given exon (Doench et al., 2014), while dCas9-KRAB knocks
down gene expression only when gRNAs are targeted to the
transcription start site (TSS) (Gilbert et al., 2014). Hence, for
this comparative study, we used the same gRNA sequence
for both CRISPRi and CRISPRn. Here, we introduced a gRNA
targeting 358 bp downstream of the
NANOG
TSS (142 bp into
exon 1 of
NANOG
) into the CRISPRi and CRISPRn clones and
selected subclones (as described in Experimental Procedures).
We then treated multiple independent subclones of CRISPRi
and CRISPRn iPSCs containing the
NANOG
gRNA-expression
vector (as indicated by mKate2 expression) with doxycycline
(Figure 2).
With CRISPRi, we found that NANOG expression was
completely lost (>99%) in multiple independent iPSC subclones
after doxycycline treatment (Figures 2A, 2C, 2E, S3A, and S3C).
However, with CRISPRn, only 60%–70%of the cells lost NANOG
expression in multiple independent subclones post-doxycycline
induction (Figures 2B, 2D, 2G, S3B, and S3D). Next, we extracted
genomic DNA from
NANOG
gRNA-containing CRISPRi and
CRISPRn iPSCs and performed sequence analysis. As expected,
we found that CRISPRi iPSCs did not harbor any mutations in the
NANOG
locus pre- or post-doxycycline treatment (Figure 2F).
However, with CRISPRn, after 12–17 days of continuous doxy-
cycline treatment, among the mutated alleles, 30%–50% of
the sequences contained in-frame INDELs at the cut site (a total
of 77 sequenced clones) (Figure 2H).
A
B
C
D
E
H
CRISPRn
CRISPRi
F
G
1
2
AAVS1 Locus
TALEN cut site
rtTA
Neo
dCas9-KRAB
KI Donor
CAG
TRE3G
SA
T2A
dCas9
p2A
mCherry
KRAB
1
2
AAVS1 Locus
TALEN cut site
rtTA
Puro
Cas9 KI
Donor
CAG
TRE3G
SA
T2A
Cas9
3XFLAG
dCas9
-KRAB
GAPDH
Dox
+ Dox
D1
Off Dox
D2 D3
Cas9
GAPDH
Dox
+ Dox
D1
Off Dox
D2 D3
Count
FLAG
FLAG
+
Count
mCherry
mCherry
+
dCas9-KRAB
DAPI
Merge
100
µ
m
Dox
dCas9-KRAB
+ Dox
DAPI
Merge
100
µ
m
Cas9
Dox
DAPI
Merge
100
µ
m
Cas9
+ Dox
DAPI
Merge
100
µ
m
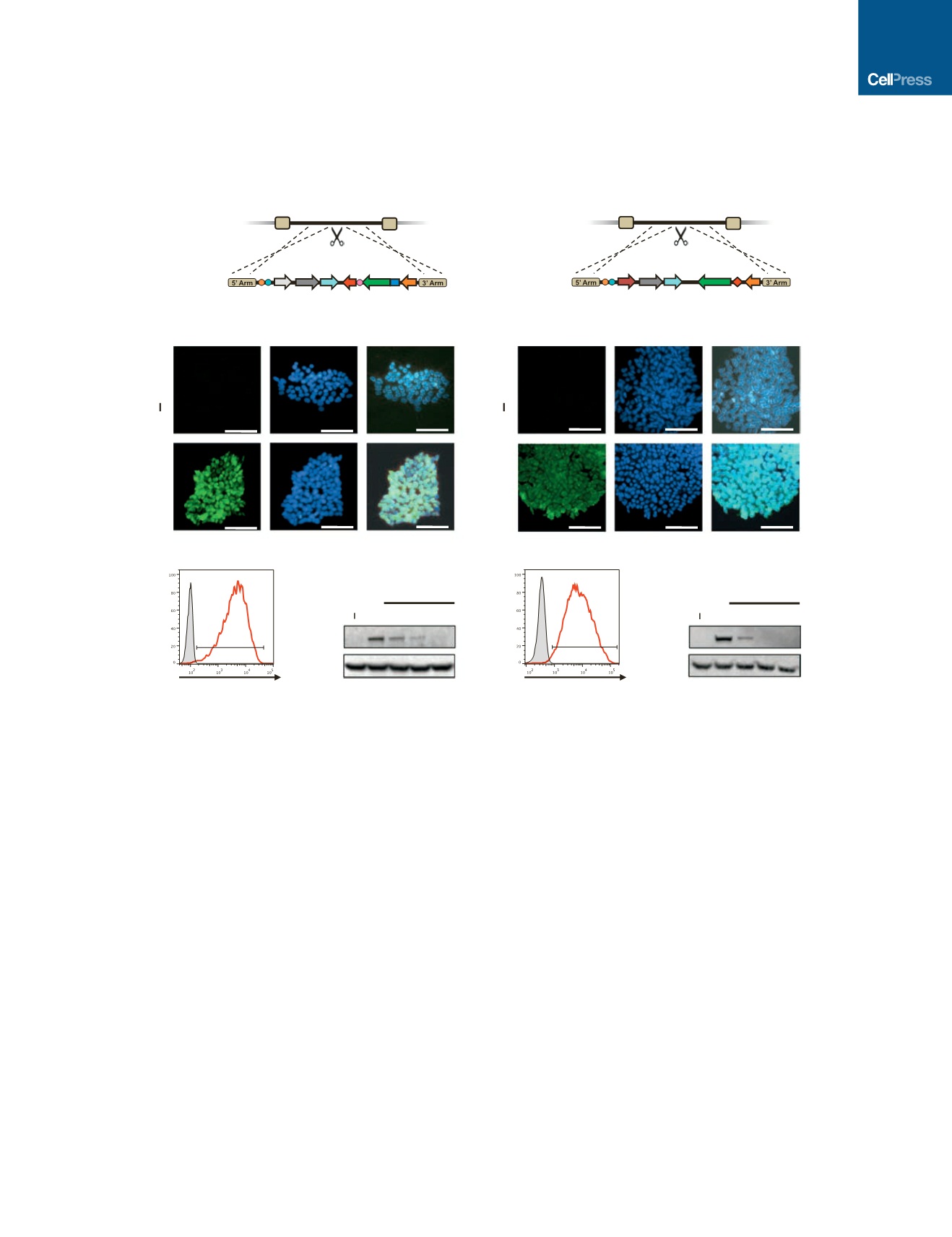
Figure 1. Generation and Characterization of Inducible CRISPRi and CRISPRn iPSCs
(A and B) Schematic overview of the strategy for TALEN-mediated targeting to the AAVS1 locus to generate the CRISPRi and CRISPRn iPSC lines. The
doxycycline-controlled reverse transcriptional activator (rtTA) is driven by a strong constitutive promoter (CAG). The third-generation doxycycline-response
element (TRE3G) drives transcription of either Cas9 (CRISPRn) or dCas9-KRAB-P2A-mCherry (CRISPRi) and is oriented in the opposite direction of the
transactivator to ensure no leaky expression without doxycycline treatment.
(C and D) Immunostaining of CRISPRi and CRISPRn colonies before and after 48 hr of doxycycline treatment with an antibody against Cas9 (green). Nuclei are
stained with DAPI (blue). All nuclei showed expression of dCas9-KRAB or Cas9 after adding doxycycline.
(E and G) Flow cytometry analysis of CRISPRi and CRISPRn iPSC lines before and after 48 hr of doxycycline treatment. Doxycycline treatment of CRISPRi and
CRISPRn produced expression of mCherry and FLAG in all cells, respectively. The doxycycline-untreated sample is plotted in gray.
(F and H) CRISPRi and CRISPRn iPSC lines were treated with doxycycline (2
m
M) for 24 hr, which was then removed to measure the protein half-life of dCas9-
KRAB and Cas9. Total protein was extracted from samples and analyzed by western blot with antibodies against Cas9 and GAPDH as a loading control. Both the
CRISPRi and CRISPRn clones express dCas9-KRAB and Cas9 at similar levels after doxycycline treatment, and the half-life of both proteins was 12 hr in iPSCs.
Scale bars, 100
m
m.
Cell Stem Cell
18
, 541–553, April 7, 2016
ª
2016 Elsevier Inc.
543


















