
muscle cells express stable dystrophin that improves membrane
stability and restores a DGC member,
b
-dystroglycan. We also
demonstrate reduced microRNA 31 (miR31) levels after the
reading frame is restored, consistent with the observations
made in BMD patients (Cacchiarelli et al., 2011). Furthermore,
we show restoration of dystrophin and
b
-dystroglycan in vivo
after engraftment of reframed hiPSC-derived skeletal muscle
cells into a mouse model of DMD. This work sets the stage for
use of reframed DMD hiPSC-derived cells or in vivo correction
strategies using CRISPR/Cas9 for direct translation to patients
with DMD.
RESULTS
DMD hiPSC Lines Are Pluripotent and Genetically Stable
We have developed several xenobiotic-free hiPSC lines derived
from wild-type and DMD patient fibroblasts using current good
manufacturing practice protocols. Each DMD hiPSC line harbors
a unique frameshifting
DMD
mutation within the exon 45–55
hotspot region. All hiPSC lines (Center for Duchenne Muscular
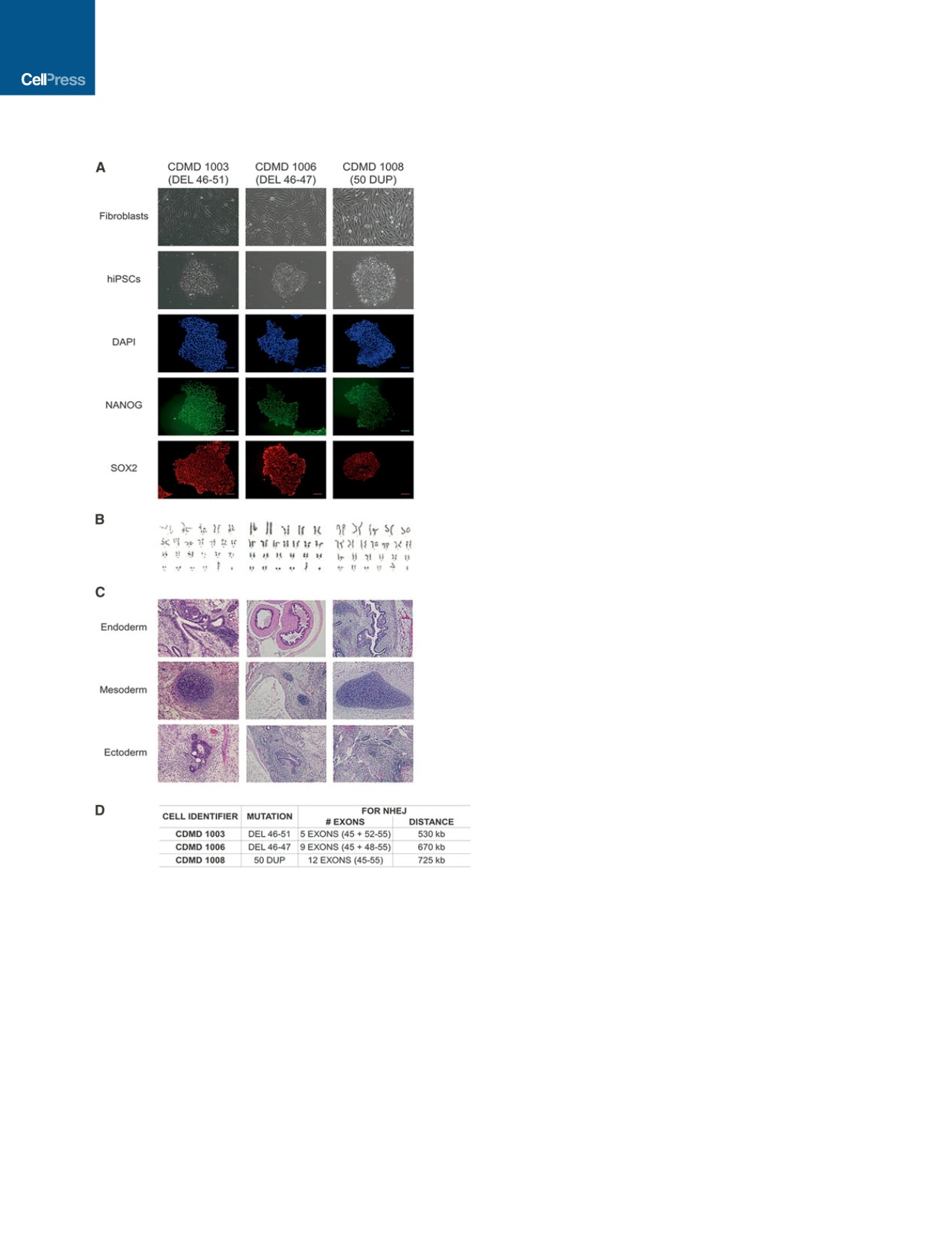
Dystrophy [CDMD] 1003, 1006, and 1008) express pluripotency
markers (NANOG and SOX2) and are karyotypically normal (Fig-
ures 1A and 1B). CDMD hiPSCs maintain pluripotency, as they
form teratomas in vivo that represent all three germ layers (Fig-
ure 1C), and each harbor unique mutations (Figure 1D).
CRISPR/Cas9-Mediated Deletion and NHEJ of up to 725
kb in the
DMD
Gene
In order to delete exons 45–55 of
DMD
, gRNAs were designed to
target introns 44 and 55. gRNA sites were chosen to only retain
500 bp of the intron next to each of the flanking exons (44 and
56). The rationale for this design is to develop gRNAs applicable
to as many patient mutations as possible and to ensure that a
small functional chimeric intron is generated. During NHEJ, the
3
0
end of intron 44 and the 5
0
end of intron 55 join to create a
1 kb chimeric intron (Figure 2A). We expect that introns gener-
ated in this manner are functional and splice correctly to create
an in-frame transcript, with exon 44 joined with exon 56.
Since hiPSCs are challenging to genetically manipulate, hu-
man embryonic kidney (HEK) 293FT cells were used to screen
five gRNAs at each intronic region. All gRNAs demonstrated in-
dividual cutting activity on Surveyor assay up to 34% (Figures
S1A and S1B). Using multiplex PCR, gRNAs transfected in pairs
were shown to effectively delete the entire 708 kb region encom-
passing exons 45–55 (Figures S1C and S1D).
In order to assess the feasibility of an exon 45–55 deletion
across different patient mutations, we applied our gRNAs to
three DMD hiPSC lines. The lines (CDMD 1003, 1006, and
1008) require 530 kb, 670 kb, or 725 kb, respectively, for suc-
cessful deletion and NHEJ of
DMD
. The gRNAs used were
shown to be active in all three lines and effectively deleted exons
45–55 (Figures S2 and S3). Transient puromycin selection of
cells nucleofected with the CRISPR plasmids improved the effi-
ciency of deletion in CDMD 1003 and 1006 hiPSCs (Figure S3D).
Clonal Reframed DMD hiPSC Lines Contain No Off-
Target Activity at Candidate Sites
Stably deleted DMD hiPSC lines were generated from CDMD
1003 and 1006 by clonal selection after nucleofection with the
gRNA pair 44C4 and 55C3 (Figures 2B and 2C) and are pluripo-
tent (Figures 2C and S4B). All reframed lines were karyotypically
normal except for one clone (CDMD 1003-81), which was found
to contain a 1q32 amplification confirmed via FISH analysis
Figure 1. CDMD hiPSCs Are Pluripotent and Genetically Stable
(A) CDMD hiPSCs were generated from DMD fibroblasts. Brightfield images
depict fibroblasts before and after reprogramming to hiPSCs. Immunocyto-
chemical staining reveals that cells express pluripotency markers NANOG
(green) and SOX2 (red). Scale bar, 100
m
m.
(B) Karyotyping of all lines is shown.
(C) CDMD hiPSCs were injected into mice to test teratoma formation in vivo.
Representative H&E stainings of the three germ layers (endoderm, mesoderm,
and ectoderm) are shown.
(D) Patient mutations for each CDMD hiPSC line are shown. In addition, the
number of exons and the approximate distance necessary for successful
NHEJ is indicated, based on comparative genomic hybridization data for the
patient’s underlying mutation size.
534
Cell Stem Cell
18
, 533–540, April 7, 2016
ª
2016 Elsevier Inc.


















