
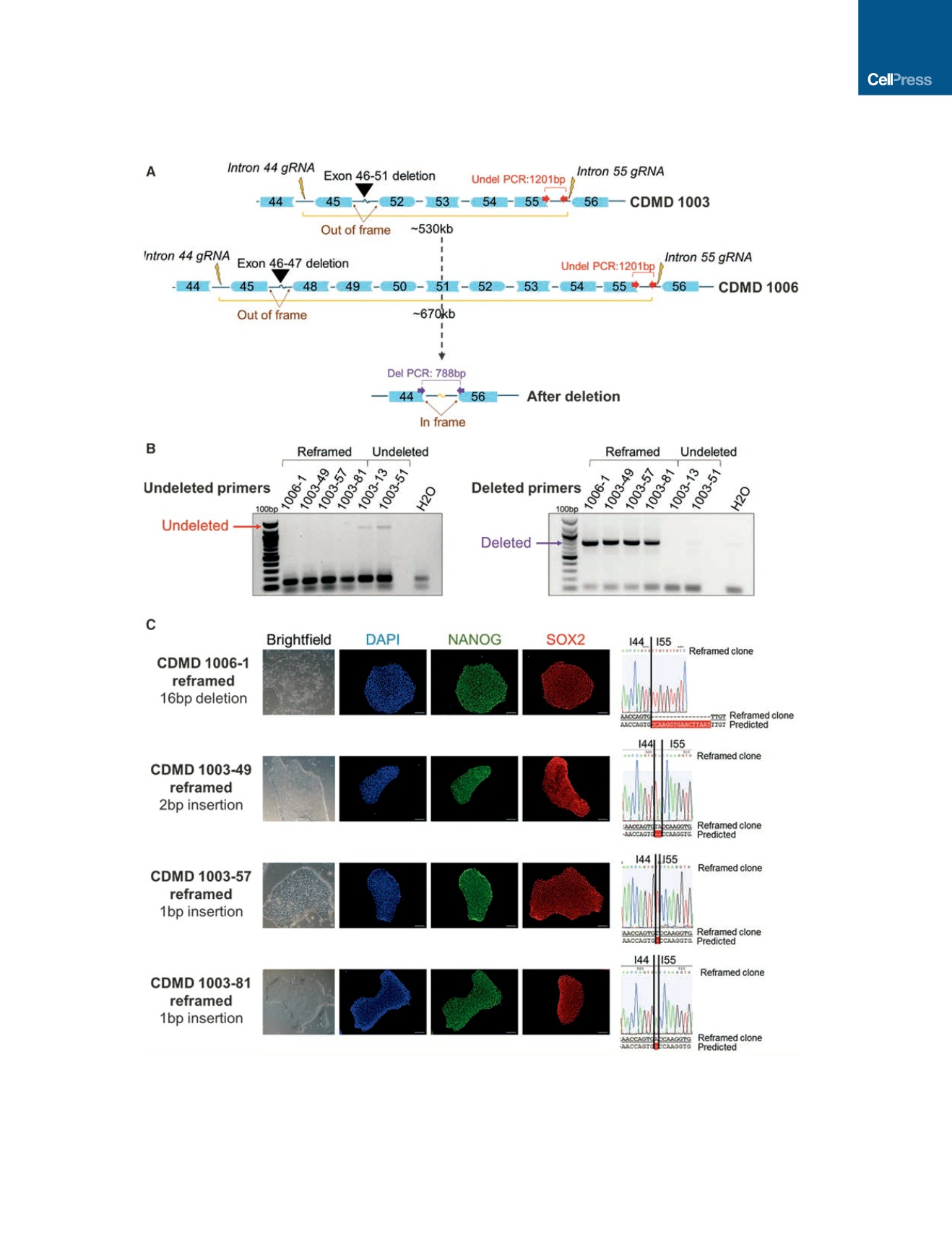
Figure 2. Generation of Stable, Pluripotent CDMD hiPSC Lines with an Exon 45–55 Deletion
(A) Shown is a cartoon (not to scale) of the region of
DMD
targeted for CRISPR/Cas9-mediated deletion using gRNAs specific to introns 44 and 55 (lightning bolts).
Successful NHEJ deletes exons 45–55 and restores the reading frame for mutations within this region. Different deletion sizes are required depending on the
patient’s underlying mutation (black arrow heads).
(B) PCR genotyping of 117 and 109 single-cell clones from parental lines CDMD 1006 and 1003, respectively, was carried out on cells nucleofected with gRNAs
44C4 and 55C3. One clone from CDMD 1006 (CDMD 1006-1) and three from CDMD 1003 (CDMD 1003-49, 1003-57, and 1003-81) were identified as stably
(legend continued on next page)
Cell Stem Cell
18
, 533–540, April 7, 2016
ª
2016 Elsevier Inc.
535


















